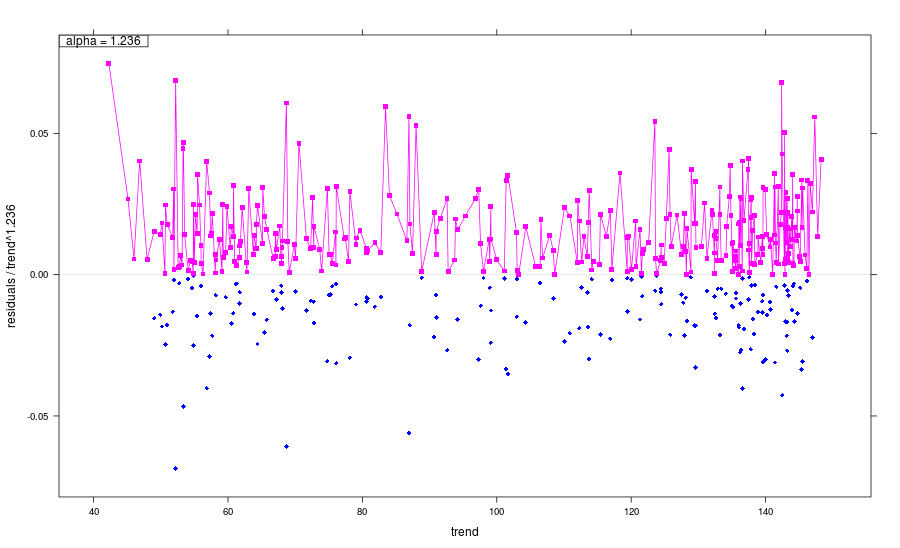
Model parameters for noise estimation
Theory about noise model used could be found here.
xlim <- c(22, 88)
ylim <- c(32, 68)
L <- c(15, 15)
file <- system.file("extdata/data", "ab16.txt", package = "BioSSA")
df <- read.emb.data(file)
bs <- BioSSA(cad ~ AP + DV, data = df, ylim = ylim, xlim = xlim, L = L)
nm <- noise.model(bs, 1:3, averaging.type = "none")
plot(plot(nm))
summary(nm)
nm <- noise.model(bs, 1:3, averaging.type = "sliding")
summary(nm)
nm <- noise.model(bs, 1:3, averaging.type = "equal")
summary(nm)
nm <- noise.model(bs, 1:3, averaging.type = "quantile")
summary(nm)
nm <- noise.model(bs, 1:3, model = "poisson")
summary(nm)
nm <- noise.model(bs, 1:3, model = "additive")
summary(nm)
nm <- noise.model(bs, 1:3, model = "multiplicative")
summary(nm)
nm <- noise.model(bs, 1:3, model = -1.2)
summary(nm)
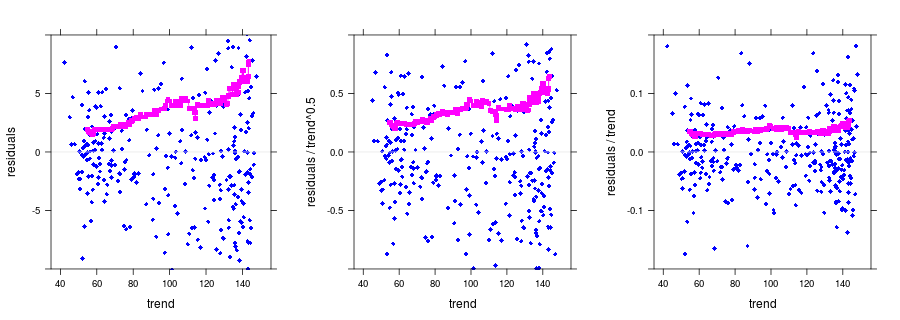
#dependence of noise on trend
good <- 3
ylim1 <- c(-10, 10)
ylim2 <- c(-1, 1)
ylim3 <- c(-0.2, 0.2)
nm.add <- noise.model(bs, groups = 1:good, model = "additive")
nm.pois <- noise.model(bs, groups = 1:good, model = "pois")
nm.mult <- noise.model(bs, groups = 1:good, model = "mult")
p1 <- plot(nm.add, ylim = ylim1, print.alpha = FALSE)
p2 <- plot(nm.pois, ylim = ylim2, print.alpha = FALSE)
p3 <- plot(nm.mult, ylim = ylim3, print.alpha = FALSE)
print(p1, split = c(1, 1, 3, 1), more = TRUE);
print(p2, split = c(2, 1, 3, 1), more = TRUE);
print(p3, split = c(3, 1, 3, 1));Produced output
Noise model:
Multiplicity: 1.236
sigma: 0.009559
Noise sd: 0.0207
Noise model:
Multiplicity: 1.349
sigma: 0.007153
Noise sd: 0.01238
Noise model:
Multiplicity: 1.268
sigma: 0.01042
Noise sd: 0.01787
Noise model:
Multiplicity: 1.36
sigma: 0.006972
Noise sd: 0.01179
Noise model:
Multiplicity: 0.5
sigma: 0.3571
Noise sd: 0.6389
Noise model:
Multiplicity: 0
sigma: 3.569
Noise sd: 7.013
Noise model:
Multiplicity: 1
sigma: 0.03572
Noise sd: 0.06115
Noise model:
Multiplicity: -1.2
sigma: 895.8
Noise sd: 2434